mVOC 4.0
Microbial volatile organic compound database
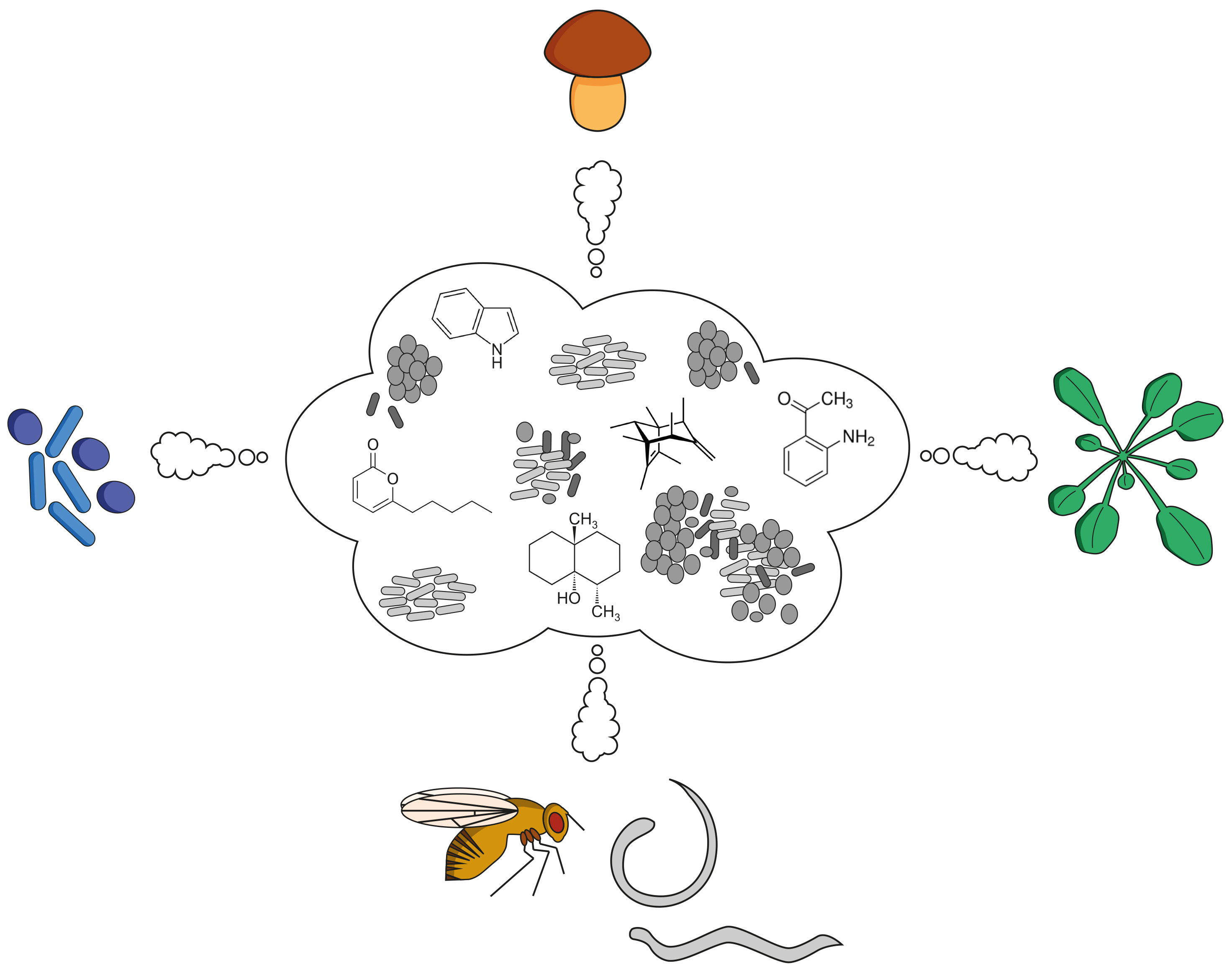
Here we present the mVOC 4.0 Database which is based on extensive literature search for microbial volatile organic compounds (mVOCs) and is an extension and improvement of the mVOC database launched in 2014 (Lemfack et al. 2014) and it's updated second version launched in 2018 (Lemfack et al. 2018). Bacteria and fungi, similar to plants and animals, emit small compounds and are an outstanding source of volatile organic compounds. Respective volatilomes are of structural complexity and diversity and possess the capability to influence neighboring organisms and communities as well as the hosts. For the first time an up-to-date data set is provided comprising the effects caused by discrete/individual mVOCs in plants, fungi, bacteria, invertebrates and vertebrates.
If you have any questions please feel free to contact us!
Please cite:
Emanuel Kemmler, Marie Chantal Lemfack, Andrean Goede, Kathleen Gallo, Serge M T Toguem, Waqar Ahmed, Iris Millberg, Saskia Preissner, Birgit Piechulla and Robert Preissner
mVOC 4.0: a database of microbial volatiles
Nucleic Acids Res. 2025 Jan 6; doi: gkae961
Search categories
The mVOC Search engine was improved. Mass Spectra search was extended. The previous search options were revised and also improved. E.g., if the user's search-string is not available in the database, the user now gets a list of similar matches. Further search options are now available via the search mVOC-button/Features/Tools-button.Basic search tools by name or chemical features were provided by mVOC Search-Site. A Mass Spectrum comparison supports compound assignment. Signature button allows the identification of fingerprint volatiles by showing the uniqueness of mVOCs or VOC profiles. KEGG pathways maps the VOCs to biosynthetic pathways. The mVOC db allows to browse for Compound Classes, Volatility, Henrys Constant and the Origin and Habitat of the microorganisms. mVOC Literature provides to search for the original literature as well as recent review articles in this research field.
Website visits
Ecological and biological functions of mVOCs
About
Recent advances highlighted the incredible complexity of the microbial populations and communities in nature, e.g. there are more than 5 x 106 bacterial species in 1 g soil, or 107 bacterial cells per cm2 leaf area, the global microbiome comprises 1030 cells and 1018 bacterial species (Farre-Armengol et al. 2016, Penuelas and Terradas 2014, Stolz 2017). In the past decade it was documented that beside diffusible compounds, microorganisms are an exceptional source of volatile organic compounds, characterized by their high vapor pressure, low boiling point and a molecular mass of below 300Da.
It is commonplace that microorganisms, bacteria and fungi, are responsible for the production of aromas of foodstuff, e.g. cheese, wine, beer, yoghurt, which have been selected for human preferences. Attention was given to mVOCs that can be used as diagnostic tools in medicine, e.g. the mVOC methyl nicotinate is emitted by Mycobacterium tuberculosis, which infects the human lung and could be used as a clinical tool for detecting tuberculosis. Furthermore mVOCs of microorganisms growing on hardware, e.g. indoor walls, are also indicators for contaminations and pollutants with potential consequences for human health (Korpi et al. 2009). Beside diverse medical applications of mVOCs new approaches in agriculture and biotechnology are also envisioned (summarized in Piechulla and Lemfack 2016).