Frequently Asked Questions

|
|
1. mVOC
In mVOC you can find information about microbial volatile compounds. For each compound you will find a 2D as well as an interactive 3D structure. Another feature of our website is a similarity search tool with which you can search for similar mVOCs for your choosen compound. KEGG pathways can be shown for several bacterial and fungal species. mVOCs as well as similar compounds to mVOCs are highlighted in these pathways. Furthermore, we provide the possibility of adding mVOCs of your own by uploading a structure.
If you use our database, please cite us. Thanks.
2. System Requirements
We recommend a recent version of Mozilla Firefox, Microsoft Internet Explorer or Google Chrome. For the usage of the ChemDoodle web component Internet Explorer 9 or newer is recommended. You find ChemDoodle implemented on the result pages for mVOC search for showing 3D structure of a mVOC, on the structure search page and on the add new mVOCs page.
3. Biological Background
3.1 What are microbial volatile organic compounds?
they are evaporating due to their low molecular weight (<300D), boiling point and high vapure pressure under ambient temperature (20oC, 101.3 kPa, defined by NIST, USA). Due to their low boiling point, such compounds can readily transform from liquid phase to gaseous phase or from solid phase to gas phase (sublimation). VOCs can travel from their source and pass through atmosphere, soil and water. Due to their low-to-moderate hydrophilicity, VOCs can dissolve in water and disperse at the air-water-interphase, excerting their infochemical effects widely, temporally and spatially.3.2 What is a functional group?
In organic chemistry, functional groups are specific groups of atoms which are responsible for characteristic chemical reactions of molecules. Chemical compounds which have the same functional group react in the same or similar way. These groups are divided into two parts due to the number of atoms:with heteroatoms and without heteroatoms. Also they affect the chemical and physical characteristics of the whole molecule. The moities are parts of a molecule including substructures of functional groups.
For example, an ester functional group is divided into an alcohol and an acyl moiety.
3.3 What physical and chemical characteristics of mVOCs are there?
Physical characters of mVOCs are:- high surface activity
- low polarity
- poor water solubility
- high lipid solubility
- high vapour pressure
Determining for mVOC charcteristics is not the chemical reactivity but a poorly polar and a highly hydrophobic part of the molecule. The polar part is also called osmophoric group. Which consists of carbonyl-, ester-, hydroxyl-, or alcoxy-moieties as well as hetero aromatic analoges. Modifications of positions of functional groups or the allylsystem can lead to a loss of volatility or modification of mVOCs.
3.4 What is an InChi?
An InCHI ( IUPAC International Chemical Identifier) consists of characters which distinctly represent a chemical substance. It is designed in a way that a single compound produces always the same identifier. Therefore, IUPAC determines a nomenclature where InCHIs are created following three steps: Normalization; Canonicalization; Serialization.There are six layers which are represented by an InCHI:
1. main layer
2. charge layer
3. stereochemical layer
4. isotopic layer
5. fixed-H-layer
6. reconnected layer
For more information about InCHIs: Visit this site.
3.5 What is a SMILES?
SMILES (Simplified Molecular Input Line Entry System) is a chemical language with which atom and bond symbols can be represented by using the ASCII characters. It is a unique string that can be used as an universal identifier for a specific chemical structure with which molecules or reactions can be symbolized.3.6 What is a fingerprint?
Fingerprints represent certain structural features of a molecule. There are two processes fingerprints are primarily used for:similarity measures like calculations of Tanimoto coeffiients, and screenings. Whereas the Tanimoto coefficient is a quantifier of similarity of two molecules. However screening is a way of eliminating molecules as candidates in a substructure search. The fingerprint algorithm examines the molecule and generates patterns of the atom. The output is a string of bits and is added to the fingerprint.
3.7 What is the tanimoto coefficient?
For similarity screening of a compound against the mVOC database fingerprints of both molecules are used. Fragments of the molecules are assigned to set bits in the 1024 bit vector fingerprint. To compare the similarity between the compounds the tanimoto coefficient is applied.
The tanimoto coefficient uses the bits set to one in both fingerprints. AB is the number of bits set to one in both molecules. A is the number of bits set to one in molecule A and B is the number of bits set to one in molecule B.
4. Data Curation
For the textming process we constructed a list of potential volatiles, a list of species and a list of search criteria. The list of potential volatiles was created as a collection of all synonyms of chemicals that where either part of the mvoc database, or where the boiling point is known to be below 250 °C.
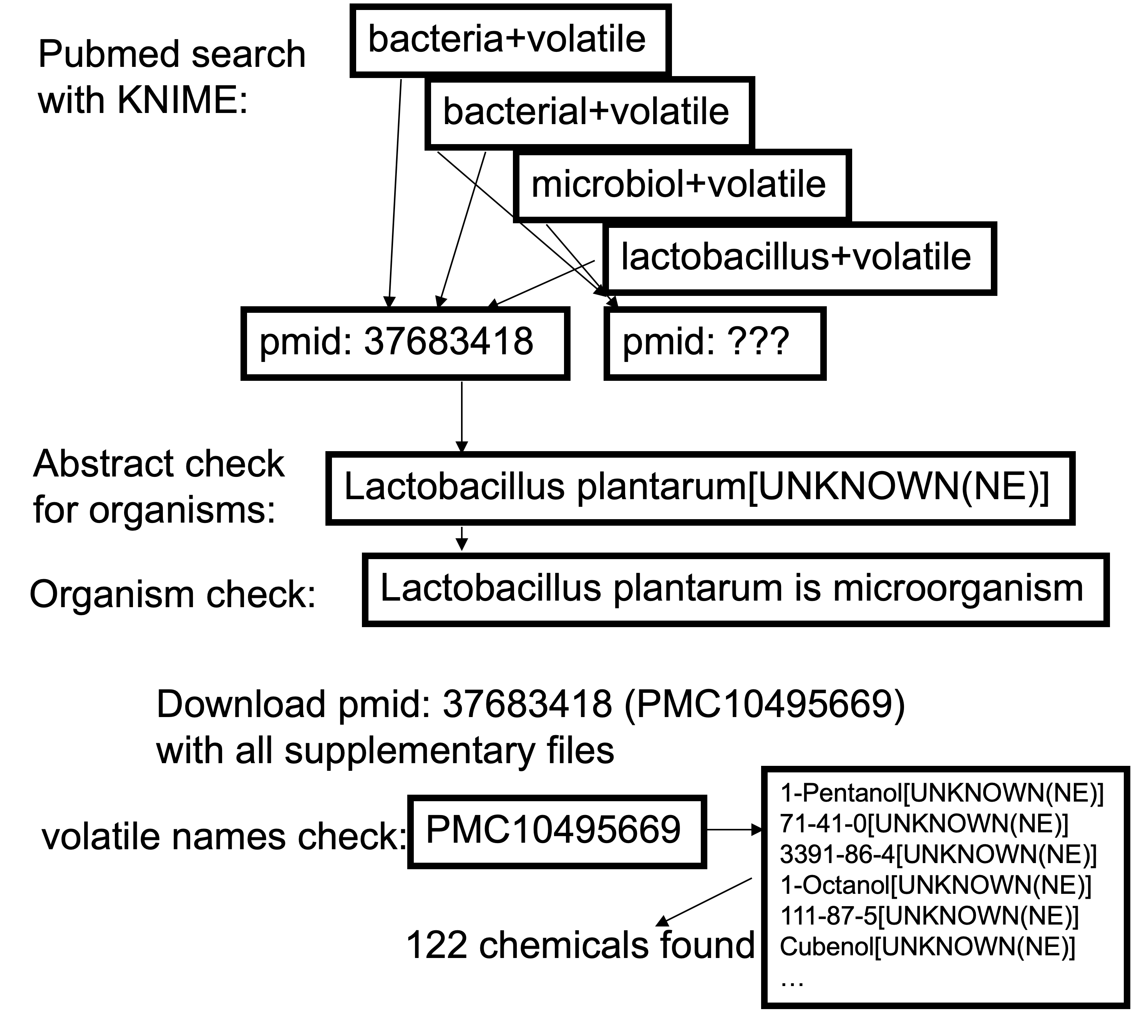
The list of search criteria was a collection of about 600 parts of species from our dabase like Acholeplasma or Actinomadura, completed by unspecific terms like bacterial, bacteria, microbiol. For the first pubmed search we defined searches for 'volatile' in combination with one of the search criteria of the list. The search was performed only in open acess publications so we were able to get the full txt including supplementary files, and only for a period of 2017 or later so there is a chance that the pdfs are in format that they can be automatically checked for text mining. The search was done with KNIME in combination with the European PubMed Central Advanced Search API.
In a first step the results of all pubmed searches were combined according to the pubmed ids. In a second step the abstracts of all these pubmed ids were checked against the list of species. A dictionary of species was created in KNIME and the search within the abstracts was conducted. The results were checked associations with microorganism. For all about 30,000 remaining publications the manuscripts including supplementary material was downloaded and checked against the list of about 450,000 synonyms of potential volatile compounds. The number of volatiles per publication was evaluated. In a last step the publications with the most mentioned volatiles were checked and volatiles produced by microorganisms were extracted.
Exemplary data curation pipeline with result table:
5. Website
5.1 Usage of the Website
The website gives you an insight of a database of mVOCS. You can search for mVOCs, search for a mVOC by its structure, browse the database, add a new mVOC or look for mVOCs in KEGG pathways at the appropriate buttons and their submenus.If you have any questions, which are not answered in the FAQs, please feel free to contact us!
5.2 What ist "Search mVOCs"?
The mVOC Search gives an assortment of different types of queries. During the search about 1,000 compounds are screened. It is possible to search for only a name, formula or the PubChem ID of a compound. Another alternative is to search for compounds which have a similar molweight, logP or the same chemical classification. A combination of the parameters allows a more specific search. The result table shows the full information with synonyms, structural properties, the compound emitters, biological effect on other organisms as well as the reference which links you to the paper.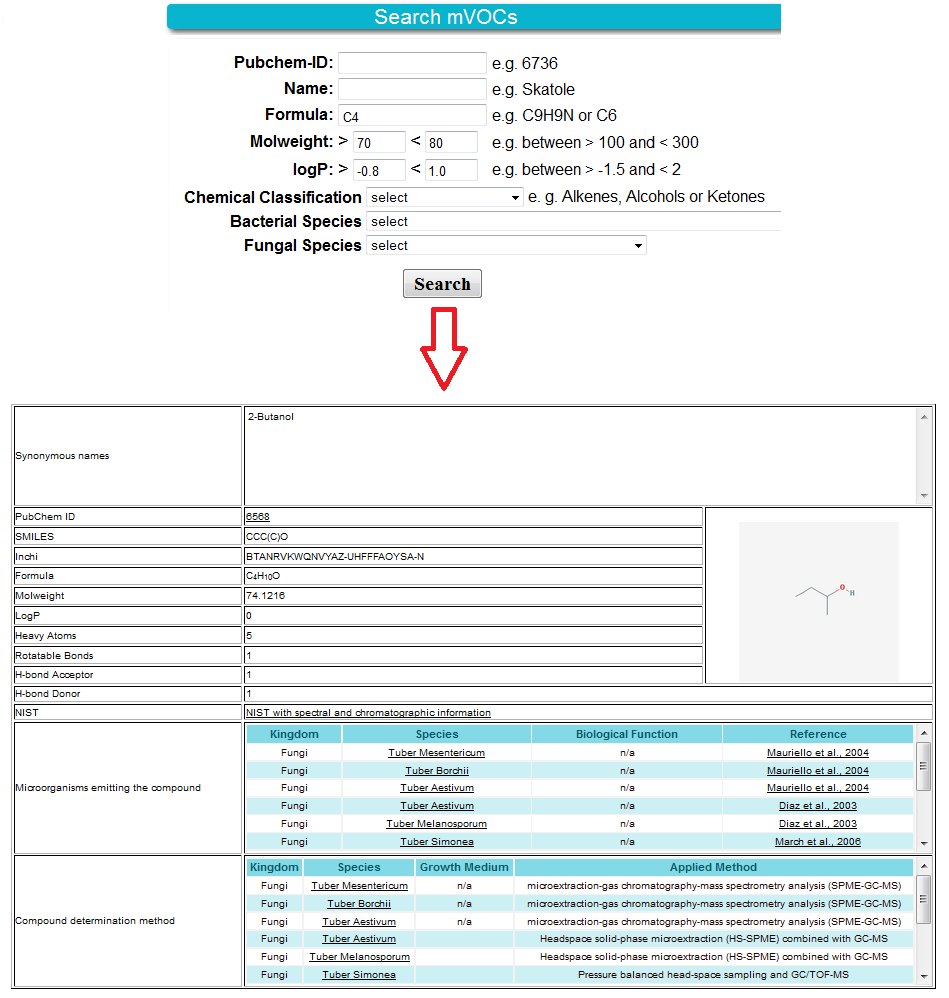
5.3 What is "Browse mVOC"?
Browse mVOC offers the possibility to browse the website by initial letters or chemical groups of all mVOCs included in mVOC database. By clicking on the respective initial letter or group a list showing mVOCs assigned to the group will appear. If you want to browse the mVOC database please click here.
5.4 What is "Signatures"?
Signature tabels of a certain species serve for the identification of signature volatiles by showing the uniqueness of mVOCs by today's scientific knowledge. In the table, compounds are plotted which are emitted by the previously chosed bacterial or fungal species. Other species that emit those compounds are plotted against them. Signature tables are useful for distinguishing between species. Based on this, new possibilities for employing diagnostic tools can be considered. Signature tables can be reached by the Signatures button. Another possibility of reaching the tables is shown below in the picture.

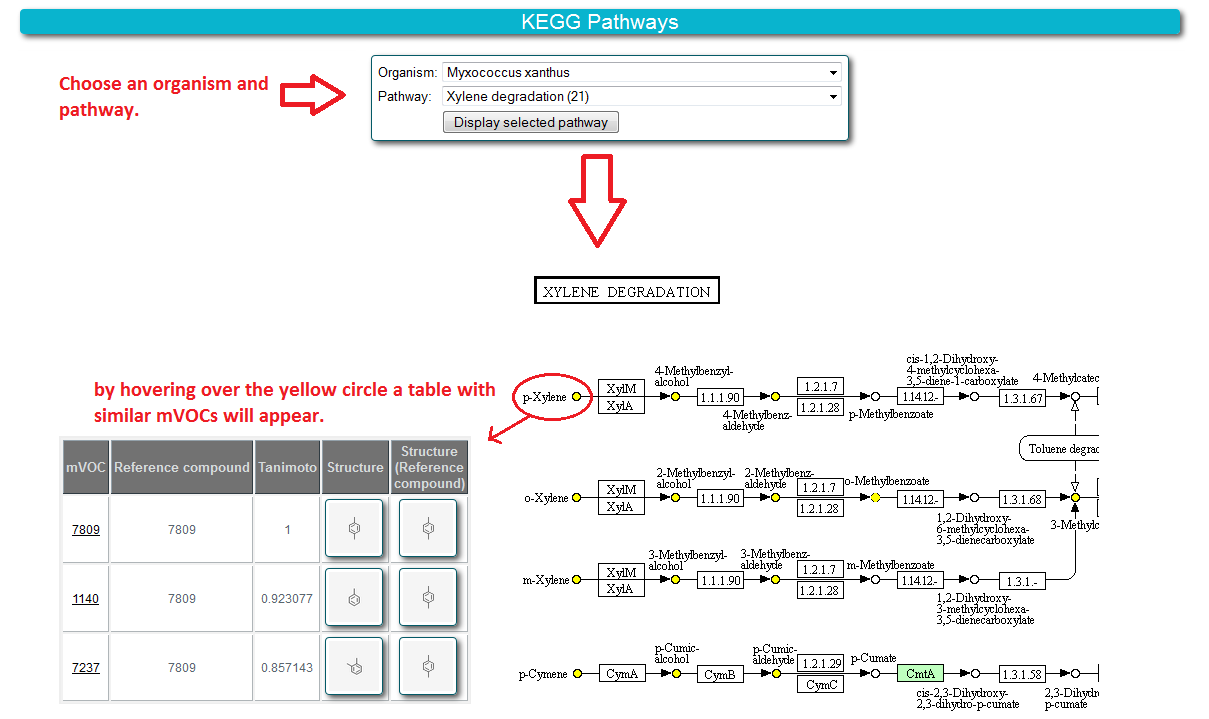
5.5 What is "Pathways"?
For biological interpretation KEGG pathways are included into the mVOC website. Compounds of mVOC database are mapped onto the KEGG pathways and highlighted in blue. Compounds similar to mVOCs contained in the database are also mapped onto the pathways and highlighted in yellow.
5.6 What is "Add a new mVOC"?
Add a new mVOC allows uploading a new mVOC in the database. The process is divided into three steps,
first: upload the structure via load a file, draw this structure or fill out the SMILES or InChI windows.
secondly: you fill out the mVOC information window providing mVOC name, organism
thirtly: you fill out the misc information window providing references and name of the user together with the e-mail address.
first: upload the structure via load a file, draw this structure or fill out the SMILES or InChI windows.
secondly: you fill out the mVOC information window providing mVOC name, organism
thirtly: you fill out the misc information window providing references and name of the user together with the e-mail address.